Example: spike_based_homeostasis
Following O. Breitwieser: “Towards a Neuromorphic Implementation of Spike-Based Expectation Maximization”
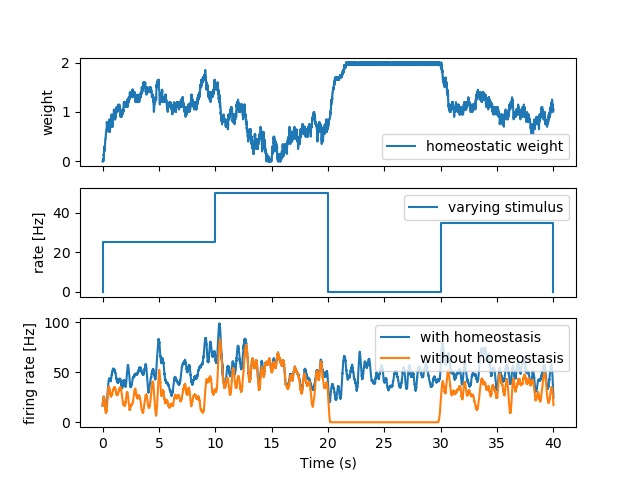
Two poisson stimuli are connected to a neuron. One with a varying rate and the other with a fixed rate. The synaptic weight from the varying rate stimulus to the neuron is fixed. The synaptic weight from the fixed rate stimulus to the neuron is plastic and tries to keep the neuron at a firing rate that is determined by the parameters of the plasticity rule.
Sebastian Schmitt, 2021
import itertools
import numpy as np
import matplotlib.pyplot as plt
from brian2 import TimedArray, PoissonGroup, NeuronGroup, Synapses, StateMonitor, PopulationRateMonitor
from brian2 import defaultclock, run
from brian2 import Hz, ms, second
# The synaptic weight from the steady stimulus is plastic
steady_stimulus = TimedArray([50]*Hz, dt=40*second)
steady_poisson = PoissonGroup(1, rates='steady_stimulus(t)')
# The synaptic weight from the varying stimulus is static
varying_stimulus = TimedArray([25*Hz, 50*Hz, 0*Hz, 35*Hz, 0*Hz], dt=10*second)
varying_poisson = PoissonGroup(1, rates='varying_stimulus(t)')
# dw_plus/dw_minus determines scales the steady stimulus rate to the target firing rate, must not be larger 1
# the magntude of dw_plus and dw_minus determines the "speed" of the homeostasis
parameters = {
'tau': 10*ms, # membrane time constant
'dw_plus': 0.05, # weight increment on pre spike
'dw_minus': 0.05, # weight increment on post spike
'w_max': 2, # maximum plastic weight
'w_initial': 0 # initial plastic weight
}
eqs = 'dv/dt = (0 - v)/tau : 1 (unless refractory)'
neuron_with_homeostasis = NeuronGroup(1, eqs,
threshold='v > 1', reset='v = -1',
method='euler', refractory=1*ms,
namespace=parameters)
neuron_without_homeostasis = NeuronGroup(1, eqs,
threshold='v > 1', reset='v = -1',
method='euler', refractory=1*ms,
namespace=parameters)
plastic_synapse = Synapses(steady_poisson, neuron_with_homeostasis,
'w : 1',
on_pre='''
v_post += w
w = clip(w + dw_plus, 0, w_max)
''',
on_post='''
w = clip(w - dw_minus, 0, w_max)
''', namespace=parameters)
plastic_synapse.connect()
plastic_synapse.w = parameters['w_initial']
non_plastic_synapse_neuron_without_homeostasis = Synapses(varying_poisson,
neuron_without_homeostasis,
'w : 1', on_pre='v_post += w')
non_plastic_synapse_neuron_without_homeostasis.connect()
non_plastic_synapse_neuron_without_homeostasis.w = 2
non_plastic_synapse_neuron = Synapses(varying_poisson, neuron_with_homeostasis,
'w : 1', on_pre='v_post += w')
non_plastic_synapse_neuron.connect()
non_plastic_synapse_neuron.w = 2
M = StateMonitor(neuron_with_homeostasis, 'v', record=True)
M2 = StateMonitor(plastic_synapse, 'w', record=True)
M_rate_neuron_with_homeostasis = PopulationRateMonitor(neuron_with_homeostasis)
M_rate_neuron_without_homeostasis = PopulationRateMonitor(neuron_without_homeostasis)
duration = 40*second
defaultclock.dt = 0.1*ms
run(duration, report='text')
fig, axes = plt.subplots(3, sharex=True)
axes[0].plot(M2.t/second, M2.w[0], label="homeostatic weight")
axes[0].set_ylabel("weight")
axes[0].legend()
# dt is in second
dts = np.arange(0., len(varying_stimulus.values)*varying_stimulus.dt, varying_stimulus.dt)
x = list(itertools.chain(*zip(dts, dts)))
y = list(itertools.chain(*zip(varying_stimulus.values/Hz, varying_stimulus.values/Hz)))
axes[1].plot(x, [0] + y[:-1], label="varying stimulus")
axes[1].set_ylabel("rate [Hz]")
axes[1].legend()
# in ms
smooth_width = 100*ms
axes[2].plot(M_rate_neuron_with_homeostasis.t/second,
M_rate_neuron_with_homeostasis.smooth_rate(width=smooth_width)/Hz,
label="with homeostasis")
axes[2].plot(M_rate_neuron_without_homeostasis.t/second,
M_rate_neuron_without_homeostasis.smooth_rate(width=smooth_width)/Hz,
label="without homeostasis")
axes[2].set_ylabel("firing rate [Hz]")
axes[2].legend()
plt.xlabel('Time (s)')
plt.show()