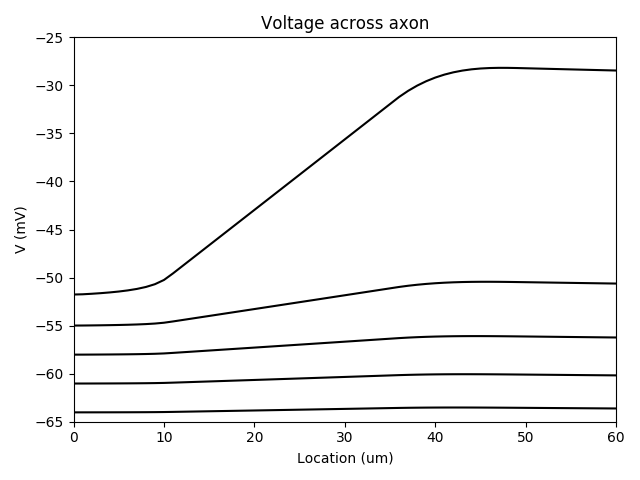
Example: Fig4
Brette R (2013). Sharpness of spike initiation in neurons explained by compartmentalization. PLoS Comp Biol, doi: 10.1371/journal.pcbi.1003338.
Fig. 4E-F. Spatial distribution of Na channels. Tapering axon near soma.
from brian2 import *
from params import *
defaultclock.dt = 0.025*ms
# Morphology
morpho = Soma(50*um) # chosen for a target Rm
# Tapering (change this for the other figure panels)
diameters = hstack([linspace(4, 1, 11), ones(290)])*um
morpho.axon = Section(diameter=diameters, length=ones(300)*um, n=300)
# Na channels
Na_start = (25 + 10)*um
Na_end = (40 + 10)*um
linear_distribution = True # True is F, False is E
duration = 500*ms
# Channels
eqs='''
Im = gL*(EL - v) + gclamp*(vc - v) + gNa*m*(ENa - v) : amp/meter**2
dm/dt = (minf - m) / taum: 1 # simplified Na channel
minf = 1 / (1 + exp((va - v) / ka)) : 1
gclamp : siemens/meter**2
gNa : siemens/meter**2
vc = EL + 50*mV * t / duration : volt (shared) # Voltage clamp with a ramping voltage command
'''
neuron = SpatialNeuron(morphology=morpho, model=eqs, Cm=Cm, Ri=Ri,
method="exponential_euler")
compartments = morpho.axon[Na_start:Na_end]
neuron.v = EL
neuron.gclamp[0] = gL*500
if linear_distribution:
profile = linspace(1, 0, len(compartments))
else:
profile = ones(len(compartments))
profile = profile / sum(profile) # normalization
neuron.gNa[compartments] = gNa_0 * profile / neuron.area[compartments]
# Monitors
mon = StateMonitor(neuron, 'v', record=True)
run(duration, report='text')
dt_per_volt = len(mon.t) / (50*mV)
for v in [-64*mV, -61*mV, -58*mV, -55*mV, -52*mV]:
plot(mon.v[:100, int(dt_per_volt * (v - EL))]/mV, 'k')
xlim(0, 50+10)
ylim(-65, -25)
ylabel('V (mV)')
xlabel('Location (um)')
title('Voltage across axon')
tight_layout()
show()