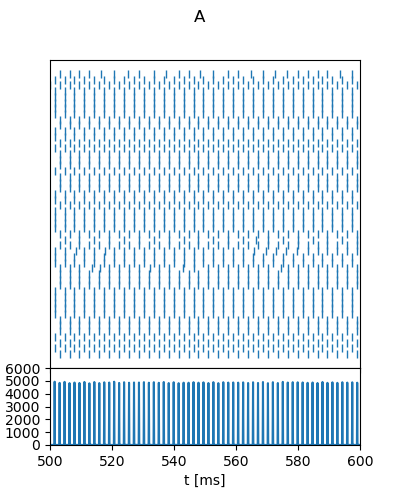
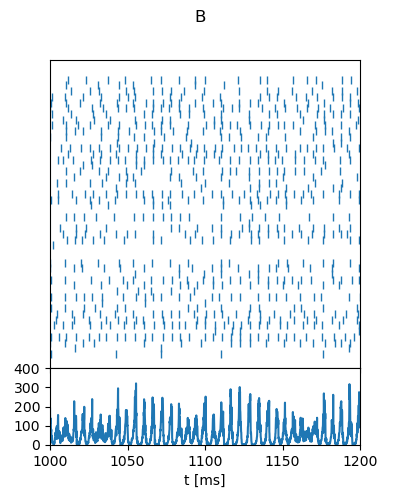
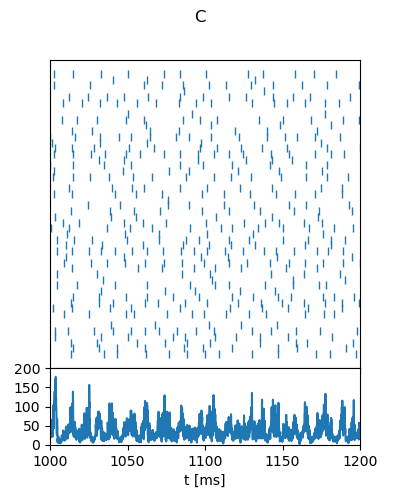
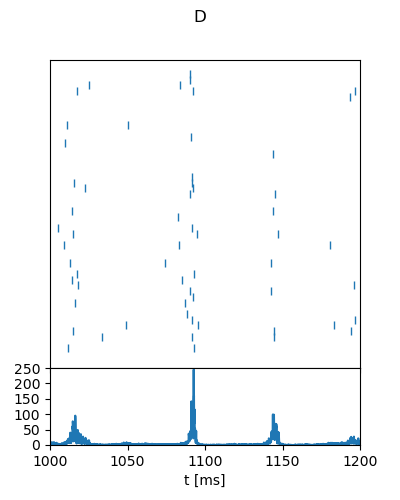
Example: Brunel_2000
Fig. 8 from:
Brunel, N. Dynamics of Sparsely Connected Networks of Excitatory and Inhibitory Spiking Neurons. J Comput Neurosci 8, 183–208 (2000). https://doi.org/10.1023/A:1008925309027
Inspired by http://neuronaldynamics.epfl.ch
Sebastian Schmitt, 2022
import random
from brian2 import *
import matplotlib.pyplot as plt
def sim(g, nu_ext_over_nu_thr, sim_time, ax_spikes, ax_rates, rate_tick_step):
"""
g -- relative inhibitory to excitatory synaptic strength
nu_ext_over_nu_thr -- ratio of external stimulus rate to threshold rate
sim_time -- simulation time
ax_spikes -- matplotlib axes to plot spikes on
ax_rates -- matplotlib axes to plot rates on
rate_tick_step -- step size for rate axis ticks
"""
# network parameters
N_E = 10000
gamma = 0.25
N_I = round(gamma * N_E)
N = N_E + N_I
epsilon = 0.1
C_E = epsilon * N_E
C_ext = C_E
# neuron parameters
tau = 20 * ms
theta = 20 * mV
V_r = 10 * mV
tau_rp = 2 * ms
# synapse parameters
J = 0.1 * mV
D = 1.5 * ms
# external stimulus
nu_thr = theta / (J * C_E * tau)
defaultclock.dt = 0.1 * ms
neurons = NeuronGroup(N,
"""
dv/dt = -v/tau : volt (unless refractory)
""",
threshold="v > theta",
reset="v = V_r",
refractory=tau_rp,
method="exact",
)
excitatory_neurons = neurons[:N_E]
inhibitory_neurons = neurons[N_E:]
exc_synapses = Synapses(excitatory_neurons, target=neurons, on_pre="v += J", delay=D)
exc_synapses.connect(p=epsilon)
inhib_synapses = Synapses(inhibitory_neurons, target=neurons, on_pre="v += -g*J", delay=D)
inhib_synapses.connect(p=epsilon)
nu_ext = nu_ext_over_nu_thr * nu_thr
external_poisson_input = PoissonInput(
target=neurons, target_var="v", N=C_ext, rate=nu_ext, weight=J
)
rate_monitor = PopulationRateMonitor(neurons)
# record from the first 50 excitatory neurons
spike_monitor = SpikeMonitor(neurons[:50])
run(sim_time, report='text')
ax_spikes.plot(spike_monitor.t / ms, spike_monitor.i, "|")
ax_rates.plot(rate_monitor.t / ms, rate_monitor.rate / Hz)
ax_spikes.set_yticks([])
ax_spikes.set_xlim(*params["t_range"])
ax_rates.set_xlim(*params["t_range"])
ax_rates.set_ylim(*params["rate_range"])
ax_rates.set_xlabel("t [ms]")
ax_rates.set_yticks(
np.arange(
params["rate_range"][0], params["rate_range"][1] + rate_tick_step, rate_tick_step
)
)
plt.subplots_adjust(hspace=0)
parameters = {
"A": {
"g": 3,
"nu_ext_over_nu_thr": 2,
"t_range": [500, 600],
"rate_range": [0, 6000],
"rate_tick_step": 1000,
},
"B": {
"g": 6,
"nu_ext_over_nu_thr": 4,
"t_range": [1000, 1200],
"rate_range": [0, 400],
"rate_tick_step": 100,
},
"C": {
"g": 5,
"nu_ext_over_nu_thr": 2,
"t_range": [1000, 1200],
"rate_range": [0, 200],
"rate_tick_step": 50,
},
"D": {
"g": 4.5,
"nu_ext_over_nu_thr": 0.9,
"t_range": [1000, 1200],
"rate_range": [0, 250],
"rate_tick_step": 50,
},
}
for panel, params in parameters.items():
fig = plt.figure(figsize=(4, 5))
fig.suptitle(panel)
gs = fig.add_gridspec(ncols=1, nrows=2, height_ratios=[4, 1])
ax_spikes, ax_rates = gs.subplots(sharex="col")
sim(
params["g"],
params["nu_ext_over_nu_thr"],
params["t_range"][1] * ms,
ax_spikes,
ax_rates,
params["rate_tick_step"],
)
plt.show()