Example: Kremer_et_al_2011_barrel_cortex
Late Emergence of the Whisker Direction Selectivity Map in the Rat Barrel Cortex. Kremer Y, Leger JF, Goodman DF, Brette R, Bourdieu L (2011). J Neurosci 31(29):10689-700.
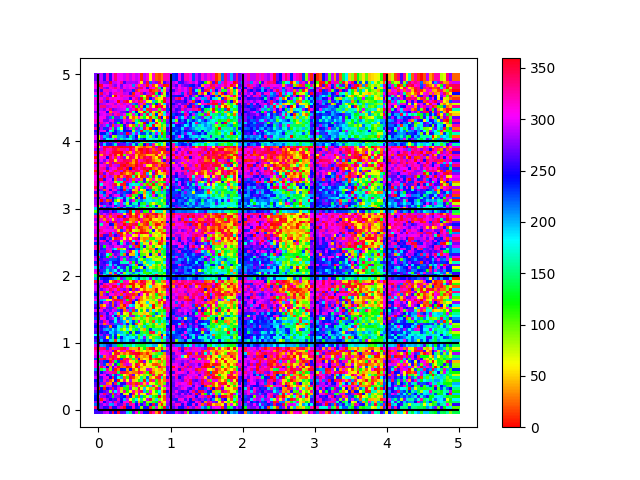
Development of direction maps with pinwheels in the barrel cortex. Whiskers are deflected with random moving bars.
N.B.: network construction can be long.
from brian2 import *
import time
t1 = time.time()
# PARAMETERS
# Neuron numbers
M4, M23exc, M23inh = 22, 25, 12 # size of each barrel (in neurons)
N4, N23exc, N23inh = M4**2, M23exc**2, M23inh**2 # neurons per barrel
barrelarraysize = 5 # Choose 3 or 4 if memory error
Nbarrels = barrelarraysize**2
# Stimulation
stim_change_time = 5*ms
Fmax = .5/stim_change_time # maximum firing rate in layer 4 (.5 spike / stimulation)
# Neuron parameters
taum, taue, taui = 10*ms, 2*ms, 25*ms
El = -70*mV
Vt, vt_inc, tauvt = -55*mV, 2*mV, 50*ms # adaptive threshold
# STDP
taup, taud = 5*ms, 25*ms
Ap, Ad= .05, -.04
# EPSPs/IPSPs
EPSP, IPSP = 1*mV, -1*mV
EPSC = EPSP * (taue/taum)**(taum/(taue-taum))
IPSC = IPSP * (taui/taum)**(taum/(taui-taum))
Ap, Ad = Ap*EPSC, Ad*EPSC
# Layer 4, models the input stimulus
eqs_layer4 = '''
rate = int(is_active)*clip(cos(direction - selectivity), 0, inf)*Fmax: Hz
is_active = abs((barrel_x + 0.5 - bar_x) * cos(direction) + (barrel_y + 0.5 - bar_y) * sin(direction)) < 0.5: boolean
barrel_x : integer # The x index of the barrel
barrel_y : integer # The y index of the barrel
selectivity : 1
# Stimulus parameters (same for all neurons)
bar_x = cos(direction)*(t - stim_start_time)/(5*ms) + stim_start_x : 1 (shared)
bar_y = sin(direction)*(t - stim_start_time)/(5*ms) + stim_start_y : 1 (shared)
direction : 1 (shared) # direction of the current stimulus
stim_start_time : second (shared) # start time of the current stimulus
stim_start_x : 1 (shared) # start position of the stimulus
stim_start_y : 1 (shared) # start position of the stimulus
'''
layer4 = NeuronGroup(N4*Nbarrels, eqs_layer4, threshold='rand() < rate*dt',
method='euler', name='layer4')
layer4.barrel_x = '(i // N4) % barrelarraysize + 0.5'
layer4.barrel_y = 'i // (barrelarraysize*N4) + 0.5'
layer4.selectivity = '(i%N4)/(1.0*N4)*2*pi' # for each barrel, selectivity between 0 and 2*pi
stimradius = (11+1)*.5
# Chose a new randomly oriented bar every 60ms
runner_code = '''
direction = rand()*2*pi
stim_start_x = barrelarraysize / 2.0 - cos(direction)*stimradius
stim_start_y = barrelarraysize / 2.0 - sin(direction)*stimradius
stim_start_time = t
'''
layer4.run_regularly(runner_code, dt=60*ms, when='start')
# Layer 2/3
# Model: IF with adaptive threshold
eqs_layer23 = '''
dv/dt=(ge+gi+El-v)/taum : volt
dge/dt=-ge/taue : volt
dgi/dt=-gi/taui : volt
dvt/dt=(Vt-vt)/tauvt : volt # adaptation
barrel_idx : integer
x : 1 # in "barrel width" units
y : 1 # in "barrel width" units
'''
layer23 = NeuronGroup(Nbarrels*(N23exc+N23inh), eqs_layer23,
threshold='v>vt', reset='v = El; vt += vt_inc',
refractory=2*ms, method='euler', name='layer23')
layer23.v = El
layer23.vt = Vt
# Subgroups for excitatory and inhibitory neurons in layer 2/3
layer23exc = layer23[:Nbarrels*N23exc]
layer23inh = layer23[Nbarrels*N23exc:]
# Layer 2/3 excitatory
# The units for x and y are the width/height of a single barrel
layer23exc.x = '(i % (barrelarraysize*M23exc)) * (1.0/M23exc)'
layer23exc.y = '(i // (barrelarraysize*M23exc)) * (1.0/M23exc)'
layer23exc.barrel_idx = 'floor(x) + floor(y)*barrelarraysize'
# Layer 2/3 inhibitory
layer23inh.x = 'i % (barrelarraysize*M23inh) * (1.0/M23inh)'
layer23inh.y = 'i // (barrelarraysize*M23inh) * (1.0/M23inh)'
layer23inh.barrel_idx = 'floor(x) + floor(y)*barrelarraysize'
print("Building synapses, please wait...")
# Feedforward connections (plastic)
feedforward = Synapses(layer4, layer23exc,
model='''w:volt
dA_source/dt = -A_source/taup : volt (event-driven)
dA_target/dt = -A_target/taud : volt (event-driven)''',
on_pre='''ge+=w
A_source += Ap
w = clip(w+A_target, 0*volt, EPSC)''',
on_post='''
A_target += Ad
w = clip(w+A_source, 0*volt, EPSC)''',
name='feedforward')
# Connect neurons in the same barrel with 50% probability
feedforward.connect('(barrel_x_pre + barrelarraysize*barrel_y_pre) == barrel_idx_post',
p=0.5)
feedforward.w = EPSC*.5
print('excitatory lateral')
# Excitatory lateral connections
recurrent_exc = Synapses(layer23exc, layer23, model='w:volt', on_pre='ge+=w',
name='recurrent_exc')
recurrent_exc.connect(p='.15*exp(-.5*(((x_pre-x_post)/.4)**2+((y_pre-y_post)/.4)**2))')
recurrent_exc.w['j<Nbarrels*N23exc'] = EPSC*.3 # excitatory->excitatory
recurrent_exc.w['j>=Nbarrels*N23exc'] = EPSC # excitatory->inhibitory
# Inhibitory lateral connections
print('inhibitory lateral')
recurrent_inh = Synapses(layer23inh, layer23exc, on_pre='gi+=IPSC',
name='recurrent_inh')
recurrent_inh.connect(p='exp(-.5*(((x_pre-x_post)/.2)**2+((y_pre-y_post)/.2)**2))')
if get_device().__class__.__name__=='RuntimeDevice':
print('Total number of connections')
print('feedforward: %d' % len(feedforward))
print('recurrent exc: %d' % len(recurrent_exc))
print('recurrent inh: %d' % len(recurrent_inh))
t2 = time.time()
print("Construction time: %.1fs" % (t2 - t1))
run(5*second, report='text')
# Calculate the preferred direction of each cell in layer23 by doing a
# vector average of the selectivity of the projecting layer4 cells, weighted
# by the synaptic weight.
_r = bincount(feedforward.j,
weights=feedforward.w * cos(feedforward.selectivity_pre)/feedforward.N_incoming,
minlength=len(layer23exc))
_i = bincount(feedforward.j,
weights=feedforward.w * sin(feedforward.selectivity_pre)/feedforward.N_incoming,
minlength=len(layer23exc))
selectivity_exc = (arctan2(_r, _i) % (2*pi))*180./pi
scatter(layer23.x[:Nbarrels*N23exc], layer23.y[:Nbarrels*N23exc],
c=selectivity_exc[:Nbarrels*N23exc],
edgecolors='none', marker='s', cmap='hsv')
vlines(np.arange(barrelarraysize), 0, barrelarraysize, 'k')
hlines(np.arange(barrelarraysize), 0, barrelarraysize, 'k')
clim(0, 360)
colorbar()
show()