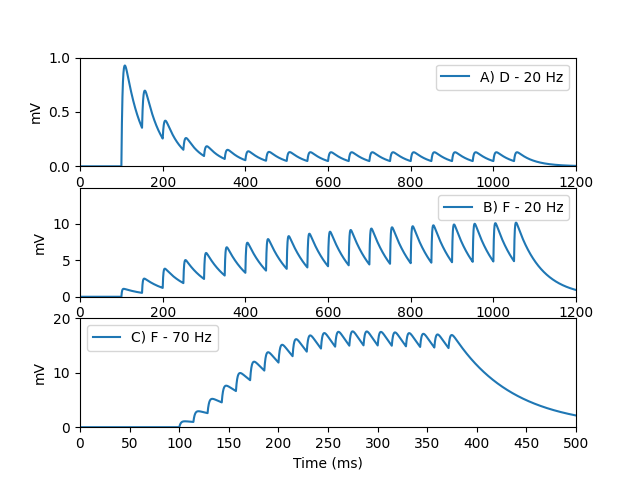
Example: Tsodyks_Pawelzik_Markram_1998
Fig. 1 from:
M. Tsodyks, K. Pawelzik, H. Markram Neural Networks with Dynamic Synapses Neural Computation 10, 821–835 (1998)
https://doi.org/10.1162/089976698300017502
Sebastian Schmitt, 2022
import numpy as np
import matplotlib.pyplot as plt
from brian2 import (
NeuronGroup,
Synapses,
SpikeGeneratorGroup,
SpikeMonitor,
StateMonitor,
)
from brian2 import ms, mV, pA, Mohm, Gohm, Hz
from brian2 import run
def get_neuron(tau_mem, R_in):
"""
tau_mem -- membrane time constant
R_in -- input resistance
"""
neuron = NeuronGroup(1,
"""
tau_mem : second
I_syn : ampere
R_in : ohm
dv/dt = -v/tau_mem + (R_in*I_syn)/tau_mem : volt
""",
method="exact")
neuron.tau_mem = tau_mem
neuron.R_in = R_in
return neuron
def get_synapses(stimulus, neuron, tau_inact, A_SE, U_SE, tau_rec, tau_facil=None):
"""
stimulus -- input stimulus
neuron -- target neuron
tau_inact -- inactivation time constant
A_SE -- absolute synaptic strength
U_SE -- utilization of synaptic efficacy
tau_rec -- recovery time constant
tau_facil -- facilitation time constant (optional)
"""
synapses_eqs = """
dx/dt = z/tau_rec : 1 (clock-driven) # recovered
dy/dt = -y/tau_inact : 1 (clock-driven) # active
A_SE : ampere
U_SE : 1
tau_inact : second
tau_rec : second
z = 1 - x - y : 1 # inactive
I_syn_post = A_SE*y : ampere (summed)
"""
if tau_facil:
synapses_eqs += """
du/dt = -u/tau_facil : 1 (clock-driven)
tau_facil : second
"""
synapses_action = """
u += U_SE*(1-u)
y += u*x # important: update y first
x += -u*x
"""
else:
synapses_action = """
y += U_SE*x # important: update y first
x += -U_SE*x
"""
synapses = Synapses(stimulus,
neuron,
model=synapses_eqs,
on_pre=synapses_action,
method="exponential_euler")
synapses.connect()
# start fully recovered
synapses.x = 1
synapses.tau_inact = tau_inact
synapses.A_SE = A_SE
synapses.U_SE = U_SE
synapses.tau_rec = tau_rec
if tau_facil:
synapses.tau_facil = tau_facil
return synapses
def get_stimulus(start, stop, frequency):
"""
start -- start time of stimulus
stop -- stop time of stimulus
frequency -- frequency of stimulus
"""
times = np.arange(start / ms, stop / ms, 1 / (frequency / Hz) * 1e3) * ms
stimulus = SpikeGeneratorGroup(1, [0] * len(times), times)
return stimulus
parameters = {
"A": {
"neuron": {"tau_mem": 40 * ms,
"R_in": 100*Mohm},
"synapse": {
"tau_inact": 3 * ms,
"A_SE": 250 * pA,
"tau_rec": 800 * ms,
"U_SE": 0.6, # 0.5 from publication does not match plot
},
"stimulus": {"start": 100 * ms,
"stop": 1100 * ms,
"frequency": 20 * Hz},
"simulation": {"duration": 1200 * ms},
"plot": {
"title": "A) D - 20 Hz",
"ylim": [0, 1],
"xlim": [0, 1200],
"xtickstep": 200,
},
},
"B": {
"neuron": {"tau_mem": 60 * ms,
"R_in": 1*Gohm},
"synapse": {
"tau_inact": 1.5 * ms,
"A_SE": 1540 * pA,
"tau_rec": 130 * ms,
"U_SE": 0.03,
"tau_facil": 530 * ms,
},
"stimulus": {"start": 100 * ms,
"stop": 1100 * ms,
"frequency": 20 * Hz},
"simulation": {"duration": 1200 * ms},
"plot": {
"title": "B) F - 20 Hz",
"ylim": [0, 14.9],
"xlim": [0, 1200],
"xtickstep": 200,
},
},
"C": {
"neuron": {"tau_mem": 60 * ms,
"R_in": 1*Gohm},
"synapse": {
"tau_inact": 1.5 * ms,
"A_SE": 1540 * pA,
"tau_rec": 130 * ms,
"U_SE": 0.03,
"tau_facil": 530 * ms,
},
"stimulus": {"start": 100 * ms,
"stop": 375 * ms,
"frequency": 70 * Hz},
"simulation": {"duration": 500 * ms},
"plot": {
"title": "C) F - 70 Hz",
"ylim": [0, 20],
"xlim": [0, 500],
"xtickstep": 50,
},
},
}
fig, axes = plt.subplots(3)
for ax, (panel, p) in zip(axes, parameters.items()):
neuron = get_neuron(**p["neuron"])
stimulus = get_stimulus(**p["stimulus"])
synapses = get_synapses(stimulus, neuron, **p["synapse"])
state_monitor_neuron = StateMonitor(neuron, ["v"], record=True)
run(p["simulation"]["duration"])
ax.plot(
state_monitor_neuron.t / ms,
state_monitor_neuron[0].v / mV,
label=p["plot"]["title"],
)
ax.set_xlim(*p["plot"]["xlim"])
ax.set_ylim(*p["plot"]["ylim"])
ax.set_ylabel("mV")
ax.set_xlabel("Time (ms)")
ax.set_xticks(
np.arange(
p["plot"]["xlim"][0],
p["plot"]["xlim"][1] + p["plot"]["xtickstep"],
p["plot"]["xtickstep"],
)
)
ax.legend()
plt.show()