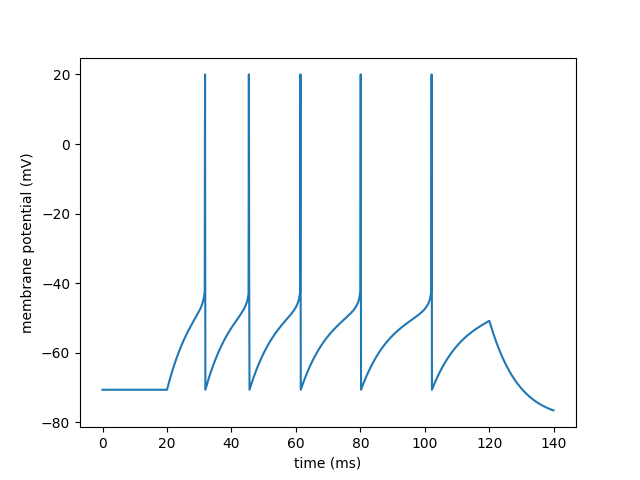
Example: Brette_Gerstner_2005
Adaptive exponential integrate-and-fire model.
http://www.scholarpedia.org/article/Adaptive_exponential_integrate-and-fire_model
Introduced in Brette R. and Gerstner W. (2005), Adaptive Exponential Integrate-and-Fire Model as an Effective Description of Neuronal Activity, J. Neurophysiol. 94: 3637 - 3642.
from brian2 import *
# Parameters
C = 281 * pF
gL = 30 * nS
taum = C / gL
EL = -70.6 * mV
VT = -50.4 * mV
DeltaT = 2 * mV
Vcut = VT + 5 * DeltaT
# Pick an electrophysiological behaviour
tauw, a, b, Vr = 144*ms, 4*nS, 0.0805*nA, -70.6*mV # Regular spiking (as in the paper)
#tauw,a,b,Vr=20*ms,4*nS,0.5*nA,VT+5*mV # Bursting
#tauw,a,b,Vr=144*ms,2*C/(144*ms),0*nA,-70.6*mV # Fast spiking
eqs = """
dvm/dt = (gL*(EL - vm) + gL*DeltaT*exp((vm - VT)/DeltaT) + I - w)/C : volt
dw/dt = (a*(vm - EL) - w)/tauw : amp
I : amp
"""
neuron = NeuronGroup(1, model=eqs, threshold='vm>Vcut',
reset="vm=Vr; w+=b", method='euler')
neuron.vm = EL
trace = StateMonitor(neuron, 'vm', record=0)
spikes = SpikeMonitor(neuron)
run(20 * ms)
neuron.I = 1*nA
run(100 * ms)
neuron.I = 0*nA
run(20 * ms)
# We draw nicer spikes
vm = trace[0].vm[:]
for t in spikes.t:
i = int(t / defaultclock.dt)
vm[i] = 20*mV
plot(trace.t / ms, vm / mV)
xlabel('time (ms)')
ylabel('membrane potential (mV)')
show()