Example: Tetzlaff_2015
Reproduces Figure 2F of
The Use of Hebbian Cell Assemblies for Nonlinear Computation by Tetzlaff C., Dasgupta S., Kulvicius T. and Wörgötter F.
Sci Rep 5, 12866 (2015). https://doi.org/10.1038/srep12866
Sebastian Schmitt, 2022
import numpy as np
import matplotlib.pyplot as plt
from brian2 import NeuronGroup, Synapses, StateMonitor, run, defaultclock, ms, second, TimedArray, seed
# random seed that gives curves similar to the ones in the publication
seed(9873487)
# neuron parameters (sigmoidal activation)
beta = 0.03
epsilon = 120
F_max = 100
F_T = 1
tau_u = 1*ms
R = 0.012
# plasticity timescales
tau_ratio = 60
# hebbian
tau_H = 3e4*ms
# synaptic scaling
tau_SS = tau_ratio * tau_H
# synaptic weights
W_max = np.sqrt(tau_ratio*(F_max**2/(F_max - F_T)))
W_ext = W_max
W_input = W_max
W_I = 0.3*W_max
# stimulus
N_units = 100
N_stim_units = 20
stim_A_units_until = N_stim_units
stim_B_units_from = N_units-N_stim_units
# connection probabilities
p_E = 0.1
p_I = 0.2
# paper uses 0.3*ms
DT = 0.5*ms
defaultclock.dt = DT
# duration of a learning trial
lt = 5000*DT
duration = 100*lt
no_input_until = 5*lt
balanced_until = duration/2
# gate balanced presentation of stimulus 1 and 2
balanced = TimedArray([lt_counter*lt < balanced_until for lt_counter in range(int(duration/lt))], dt=lt)
# function used for stimulus (typo in paper, +1 is not part of the argument of sin)
stim_func = TimedArray([100*(np.sin(0.1*(i+1))+1) for i in range(int(duration/DT))], dt=DT)
# gate learning phase of either stimulus 1 or 2
learning_phase = TimedArray([i%10 > 3 for i in range(int(duration/(0.1*lt)))], dt=0.1*lt)
# if not balanced present stimulus A three times more often than stimulus B
stim_A_gate = TimedArray([lt_counter % 2 == 0 if balanced(lt_counter*lt) else lt_counter % 4 in [0,1,2]
for lt_counter in range(int(duration/lt))], dt=lt)
stim_B_gate = TimedArray([lt_counter % 2 == 1 if balanced(lt_counter*lt) else lt_counter % 4 == 3
for lt_counter in range(int(duration/lt))], dt=lt)
# noise is applied also during stimulation
neurons = NeuronGroup(N_units,
"""
F = F_max/(1+exp(beta*(epsilon-u))) : 1
du/dt = (-u + R*(I_E - I_I + W_input*(I_stim_A + I_stim_B)))/tau_u + R*W_ext*20*sqrt((DT/ms)/ms)*xi: 1
I_E : 1
I_I : 1
index : 1 (constant)
stim_units_A = index < stim_A_units_until : boolean
stim_units_B = index >= (stim_B_units_from) : boolean
I_stim_A = learning_phase(t)*int(stim_units_A)*stim_A_gate(t)*stim_func(t) : 1
I_stim_B = learning_phase(t)*int(stim_units_B)*stim_B_gate(t)*stim_func(t) : 1
""",
method = "euler")
neurons.index = range(len(neurons))
# excitatory connections with Hebbian plasticity and synaptic scaling
synapses_E = Synapses(neurons, neurons,
"""
dw/dt = 1/tau_H*F_pre*F_post + 1/tau_SS*(F_T - F_post)*w**2 : 1 (clock-driven)
I_E_post = w*F_pre : 1 (summed)
""",
method="euler"
)
# do not connect between the two populations of stimulated neurons
synapses_E.connect(p=p_E, condition="((j > stim_A_units_until and i >= stim_B_units_from) or (j < stim_B_units_from and i < stim_A_units_until))"
"or ((i > stim_A_units_until and i < stim_B_units_from) and (j > stim_A_units_until and j < stim_B_units_from))")
# fixed weight inhibitory connections
synapses_I = Synapses(neurons, neurons,
"""
w : 1
I_I_post = w*F_pre : 1 (summed)
"""
)
synapses_I.connect(p=p_I)
synapses_I.w = W_I
statemon_neurons = StateMonitor(neurons, ["F", "I_stim_A", "I_stim_B"], record=True, dt=100*defaultclock.dt)
statemon_synapses_E = StateMonitor(synapses_E, "w", record=True, dt=100*defaultclock.dt)
statemon_synapses_for_assembly_analysis = StateMonitor(synapses_E, "w", record=True, dt=lt)
run(duration, report="text")
# threshold saying that synaptic efficacies larger than theta are
# 'strong' and others are 'weak'
theta = 0.5*W_max
in_assembly_A = []
in_assembly_B = []
# traverse through the graph following 'strong' synapses
def go(W, source, units_in_assembly):
units_in_assembly.add(source)
# check all possible targets
for target in range(N_units):
w = W[source][target]
if w > theta:
W[source][target] = 0
go(W, target, units_in_assembly)
# for each learning trial
for ws in statemon_synapses_for_assembly_analysis.w.T:
# construct a full weight matrix
W = np.full((N_units, N_units), np.nan)
W[synapses_E.i[:], synapses_E.j[:]] = ws
for in_assembly, stim_units in zip([in_assembly_A, in_assembly_B],
[range(stim_A_units_until),
range(stim_B_units_from, N_units)]):
units_in_assembly = set()
# start with units that are stimulated
for stim_unit in stim_units:
go(W, stim_unit, units_in_assembly)
in_assembly.append(len(units_in_assembly))
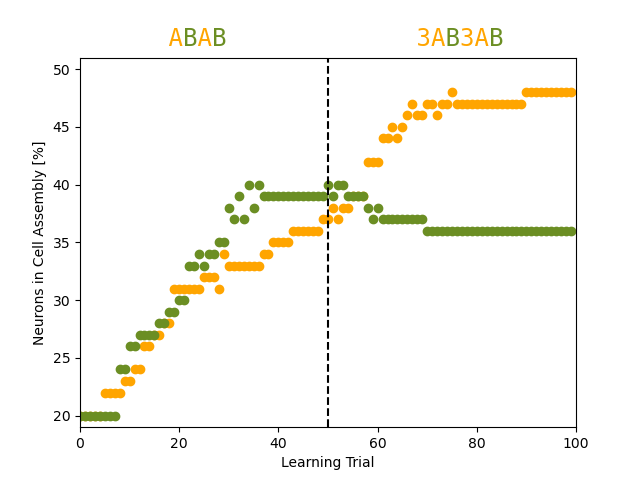
# competitive development of the two competing cell assemblies A and B as a function of the input protocol
fig, ax = plt.subplots()
ax.plot(in_assembly_A, linestyle="None", marker='o', color='orange', label="A")
ax.plot(in_assembly_B, linestyle="None", marker='o', color='olivedrab', label="B")
ax.set_ylim(19, 51)
ax.set_xlim(0, 100)
ax.set_ylabel("Neurons in Cell Assembly [%]")
ax.set_xlabel("Learning Trial")
ax.axvline(balanced_until/lt, linestyle='dashed', color='k')
ax.text(15, 52, " A A", color='orange', fontfamily="monospace", fontsize="xx-large")
ax.text(15, 52, " B B", color='olivedrab', fontfamily="monospace", fontsize="xx-large")
ax.text(65, 52, " 3A 3A", color='orange', fontfamily="monospace", fontsize="xx-large")
ax.text(65, 52, " B B", color='olivedrab', fontfamily="monospace", fontsize="xx-large")
plt.show()
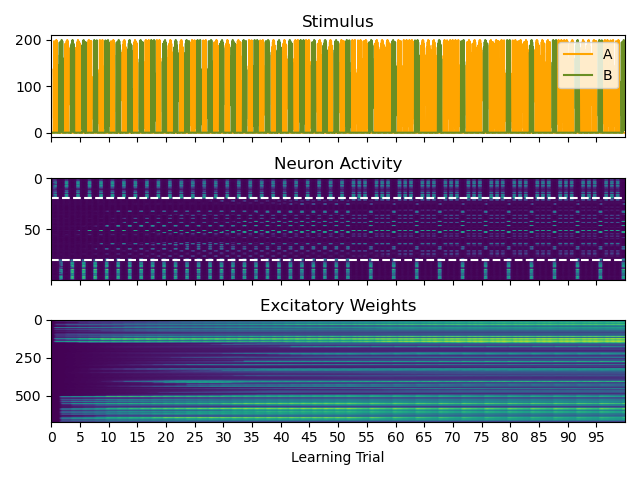
# stimulus, neuronal activity and excitatory weights as function of time
fig, axes = plt.subplots(3, sharex=True)
axes[0].plot(statemon_neurons.I_stim_A[0], label="A", color='orange')
axes[0].plot(statemon_neurons.I_stim_B[-1], label="B", color='olivedrab')
axes[0].legend(loc="upper right")
axes[0].set_title("Stimulus")
axes[1].imshow(statemon_neurons.F, aspect='auto')
axes[1].set_title("Neuron Activity")
axes[1].axhline(stim_A_units_until, linestyle='dashed', color='white')
axes[1].axhline(stim_B_units_from, linestyle='dashed', color='white')
axes[2].imshow(statemon_synapses_E.w, aspect='auto')
axes[2].set_title("Excitatory Weights")
axes[2].set_xticks(range(0, 5000, 250))
axes[2].set_xticklabels(f"{i}" for i in range(0, 100, 5))
axes[2].set_xlabel("Learning Trial")
axes[2].set_xlim(0, 5000)
fig.tight_layout()
plt.show()