Example: Rossant_et_al_2011bis
Distributed synchrony example
Fig. 14 from:
Rossant C, Leijon S, Magnusson AK, Brette R (2011). “Sensitivity of noisy neurons to coincident inputs”. Journal of Neuroscience, 31(47).
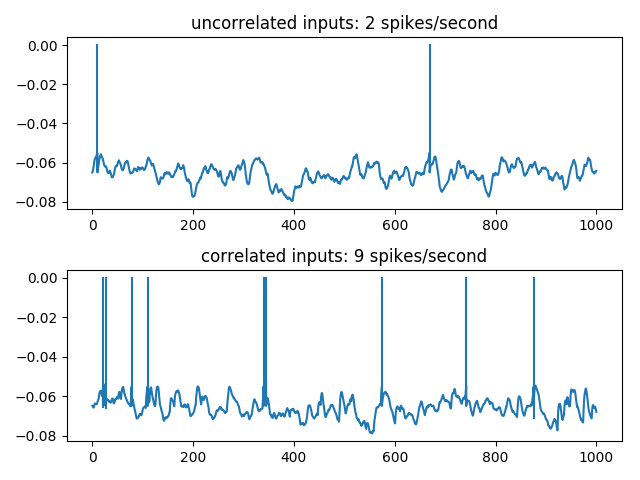
5000 independent E/I Poisson inputs are injected into a leaky integrate-and-fire neuron. Synchronous events, following an independent Poisson process at 40 Hz, are considered, where 15 E Poisson spikes are randomly shifted to be synchronous at those events. The output firing rate is then significantly higher, showing that the spike timing of less than 1% of the excitatory synapses have an important impact on the postsynaptic firing.
from brian2 import *
# neuron parameters
theta = -55*mV
El = -65*mV
vmean = -65*mV
taum = 5*ms
taue = 3*ms
taui = 10*ms
eqs = Equations("""
dv/dt = (ge+gi-(v-El))/taum : volt
dge/dt = -ge/taue : volt
dgi/dt = -gi/taui : volt
""")
# input parameters
p = 15
ne = 4000
ni = 1000
lambdac = 40*Hz
lambdae = lambdai = 1*Hz
# synapse parameters
we = .5*mV/(taum/taue)**(taum/(taue-taum))
wi = (vmean-El-lambdae*ne*we*taue)/(lambdae*ni*taui)
# NeuronGroup definition
group = NeuronGroup(N=2, model=eqs, reset='v = El',
threshold='v>theta',
refractory=5*ms, method='exact')
group.v = El
group.ge = group.gi = 0
# independent E/I Poisson inputs
p1 = PoissonInput(group[0:1], 'ge', N=ne, rate=lambdae, weight=we)
p2 = PoissonInput(group[0:1], 'gi', N=ni, rate=lambdai, weight=wi)
# independent E/I Poisson inputs + synchronous E events
p3 = PoissonInput(group[1:], 'ge', N=ne, rate=lambdae-(p*1.0/ne)*lambdac, weight=we)
p4 = PoissonInput(group[1:], 'gi', N=ni, rate=lambdai, weight=wi)
p5 = PoissonInput(group[1:], 'ge', N=1, rate=lambdac, weight=p*we)
# run the simulation
M = SpikeMonitor(group)
SM = StateMonitor(group, 'v', record=True)
BrianLogger.log_level_info()
run(1*second)
# plot trace and spikes
for i in [0, 1]:
spikes = (M.t[M.i == i] - defaultclock.dt)/ms
val = SM[i].v
subplot(2, 1, i+1)
plot(SM.t/ms, val)
plot(tile(spikes, (2, 1)),
vstack((val[array(spikes, dtype=int)],
zeros(len(spikes)))), 'C0')
title("%s: %d spikes/second" % (["uncorrelated inputs", "correlated inputs"][i],
M.count[i]))
tight_layout()
show()