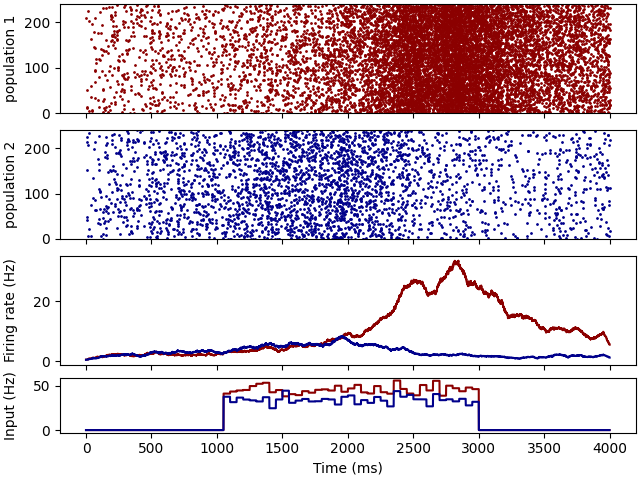
Example: Wang_2002
Decision network as in:
Wang, X.-J. Probabilistic decision making by slow reverberation in cortical circuits. Neuron, 2002, 36, 955-968.
Authors: Klaus Wimmer (kwimmer@crm.cat) and Marcel Stimberg
from brian2 import *
# -----------------------------------------------------------------------------------------------
# Set up the simulation
# -----------------------------------------------------------------------------------------------
# Stimulus and simulation parameters
coh = 12.8 # coherence of random dots
sigma = 4.0 * Hz # standard deviation of stimulus input
mu0 = 40.0 * Hz # stimulus input at zero coherence
mu1 = 40.0 * Hz # selective stimulus input at highest coherence
stim_interval = 50.0 * ms # stimulus changes every 50 ms
stim_on = 1000 * ms # stimulus onset
stim_off = 3000 * ms # stimulus offset
runtime = 4000 * ms # total simulation time
# External noise inputs
N_ext = 1000 # number of external Poisson neurons
rate_ext_E = 2400 * Hz / N_ext # external Poisson rate for excitatory population
rate_ext_I = 2400 * Hz / N_ext # external Poisson rate for inhibitory population
# Network parameters
N = 2000 # number of neurons
f_inh = 0.2 # fraction of inhibitory neurons
NE = int(N * (1.0 - f_inh)) # number of excitatory neurons (1600)
NI = int(N * f_inh) # number of inhibitory neurons (400)
fE = 0.15 # coding fraction
subN = int(fE * NE) # number of neurons in decision pools (240)
# Neuron parameters
El = -70.0 * mV # resting potential
Vt = -50.0 * mV # firing threshold
Vr = -55.0 * mV # reset potential
CmE = 0.5 * nF # membrane capacitance for pyramidal cells (excitatory neurons)
CmI = 0.2 * nF # membrane capacitance for interneurons (inhibitory neurons)
gLeakE = 25.0 * nS # membrane leak conductance of excitatory neurons
gLeakI = 20.0 * nS # membrane leak conductance of inhibitory neurons
refE = 2.0 * ms # refractory periodof excitatory neurons
refI = 1.0 * ms # refractory period of inhibitory neurons
# Synapse parameters
V_E = 0. * mV # reversal potential for excitatory synapses
V_I = -70. * mV # reversal potential for inhibitory synapses
tau_AMPA = 2.0 * ms # AMPA synapse decay
tau_NMDA_rise = 2.0 * ms # NMDA synapse rise
tau_NMDA_decay = 100.0 * ms # NMDA synapse decay
tau_GABA = 5.0 * ms # GABA synapse decay
alpha = 0.5 * kHz # saturation of NMDA channels at high presynaptic firing rates
C = 1 * mmole # extracellular magnesium concentration
# Synaptic conductances
gextE = 2.1 * nS # external -> excitatory neurons (AMPA)
gextI = 1.62 * nS # external -> inhibitory neurons (AMPA)
gEEA = 0.05 * nS / NE * 1600 # excitatory -> excitatory neurons (AMPA)
gEIA = 0.04 * nS / NE * 1600 # excitatory -> inhibitory neurons (AMPA)
gEEN = 0.165 * nS / NE * 1600 # excitatory -> excitatory neurons (NMDA)
gEIN = 0.13 * nS / NE * 1600 # excitatory -> inhibitory neurons (NMDA)
gIE = 1.3 * nS / NI * 400 # inhibitory -> excitatory neurons (GABA)
gII = 1.0 * nS / NI * 400 # inhibitory -> inhibitory neurons (GABA)
# Synaptic footprints
Jp = 1.7 # relative synaptic strength inside a selective population (1.0: no potentiation))
Jm = 1.0 - fE * (Jp - 1.0) / (1.0 - fE)
# Neuron equations
# Note the "(unless refractory)" statement serves to clamp the membrane voltage during the refractory period;
# otherwise the membrane potential continues to be integrated but no spikes are emitted.
eqsE = """
label : integer (constant) # label for decision encoding populations
dV/dt = (- gLeakE * (V - El) - I_AMPA - I_NMDA - I_GABA - I_AMPA_ext + I_input) / CmE : volt (unless refractory)
I_AMPA = s_AMPA * (V - V_E) : amp
ds_AMPA / dt = - s_AMPA / tau_AMPA : siemens
I_NMDA = gEEN * s_NMDA_tot * (V - V_E) / ( 1 + exp(-0.062 * V/mvolt) * (C/mmole / 3.57) ) : amp
s_NMDA_tot : 1
I_GABA = s_GABA * (V - V_I) : amp
ds_GABA / dt = - s_GABA / tau_GABA : siemens
I_AMPA_ext = s_AMPA_ext * (V - V_E) : amp
ds_AMPA_ext / dt = - s_AMPA_ext / tau_AMPA : siemens
I_input : amp
ds_NMDA / dt = - s_NMDA / tau_NMDA_decay + alpha * x * (1 - s_NMDA) : 1
dx / dt = - x / tau_NMDA_rise : 1
"""
eqsI = """
dV/dt = (- gLeakI * (V - El) - I_AMPA - I_NMDA - I_GABA - I_AMPA_ext) / CmI : volt (unless refractory)
I_AMPA = s_AMPA * (V - V_E) : amp
ds_AMPA / dt = - s_AMPA / tau_AMPA : siemens
I_NMDA = gEIN * s_NMDA_tot * (V - V_E) / ( 1 + exp(-0.062 * V/mvolt) * (C/mmole / 3.57) ): amp
s_NMDA_tot : 1
I_GABA = s_GABA * (V - V_I) : amp
ds_GABA / dt = - s_GABA / tau_GABA : siemens
I_AMPA_ext = s_AMPA_ext * (V - V_E) : amp
ds_AMPA_ext / dt = - s_AMPA_ext / tau_AMPA : siemens
"""
# Neuron populations
popE = NeuronGroup(NE, model=eqsE, threshold='V > Vt', reset='V = Vr', refractory=refE, method='euler', name='popE')
popI = NeuronGroup(NI, model=eqsI, threshold='V > Vt', reset='V = Vr', refractory=refI, method='euler', name='popI')
popE1 = popE[:subN]
popE2 = popE[subN:2 * subN]
popE3 = popE[2 * subN:]
popE1.label = 0
popE2.label = 1
popE3.label = 2
# Recurrent excitatory -> excitatory connections mediated by AMPA receptors
C_EE_AMPA = Synapses(popE, popE, 'w : siemens', on_pre='s_AMPA += w', delay=0.5 * ms, method='euler', name='C_EE_AMPA')
C_EE_AMPA.connect()
C_EE_AMPA.w[:] = gEEA
C_EE_AMPA.w["label_pre == label_post and label_pre < 2"] = gEEA*Jp
C_EE_AMPA.w["label_pre != label_post and label_post < 2"] = gEEA*Jm
# Note that this produces the following structure of excitatory connections:
#
# | from E1 from E2 from E3
# ---------------------------------
# to E1 | Jp Jm Jm
# to E2 | Jm Jp Jm
# to E3 | 1 1 1
# Recurrent excitatory -> inhibitory connections mediated by AMPA receptors
C_EI_AMPA = Synapses(popE, popI, on_pre='s_AMPA += gEIA', delay=0.5 * ms, method='euler', name='C_EI_AMPA')
C_EI_AMPA.connect()
# Recurrent excitatory -> excitatory connections mediated by NMDA receptors
C_EE_NMDA = Synapses(popE, popE, on_pre='x_pre += 1', delay=0.5 * ms, method='euler', name='C_EE_NMDA')
C_EE_NMDA.connect(j='i')
# Dummy population to store the summed activity of the three populations
NMDA_sum_group = NeuronGroup(3, 's : 1', name='NMDA_sum_group')
# Sum the activity according to the subpopulation labels
NMDA_sum = Synapses(popE, NMDA_sum_group, 's_post = s_NMDA_pre : 1 (summed)', name='NMDA_sum')
NMDA_sum.connect(j='label_pre')
# Propagate the summed activity to the NMDA synapses
NMDA_set_total_E = Synapses(NMDA_sum_group, popE,
'''w : 1 (constant)
s_NMDA_tot_post = w*s_pre : 1 (summed)''', name='NMDA_set_total_E')
NMDA_set_total_E.connect()
NMDA_set_total_E.w = 1
NMDA_set_total_E.w["i == label_post and label_post < 2"] = Jp
NMDA_set_total_E.w["i != label_post and label_post < 2"] = Jm
# Recurrent excitatory -> inhibitory connections mediated by NMDA receptors
NMDA_set_total_I = Synapses(NMDA_sum_group, popI,
'''s_NMDA_tot_post = s_pre : 1 (summed)''', name='NMDA_set_total_I')
NMDA_set_total_I.connect()
# Recurrent inhibitory -> excitatory connections mediated by GABA receptors
C_IE = Synapses(popI, popE, on_pre='s_GABA += gIE', delay=0.5 * ms, method='euler', name='C_IE')
C_IE.connect()
# Recurrent inhibitory -> inhibitory connections mediated by GABA receptors
C_II = Synapses(popI, popI, on_pre='s_GABA += gII', delay=0.5 * ms, method='euler', name='C_II')
C_II.connect()
# External inputs (fixed background firing rates)
extinputE = PoissonInput(popE, 's_AMPA_ext', N_ext, rate_ext_E, gextE)
extinputI = PoissonInput(popI, 's_AMPA_ext', N_ext, rate_ext_I, gextI)
# Stimulus input (updated every 50ms)
stiminputE1 = PoissonGroup(subN, rates=0*Hz, name='stiminputE1')
stiminputE2 = PoissonGroup(subN, rates=0*Hz, name='stiminputE2')
stiminputE1.run_regularly("rates = int(t > stim_on and t < stim_off) * (mu0 + coh / 100.0 * mu1 + sigma*randn())", dt=stim_interval)
stiminputE2.run_regularly("rates = int(t > stim_on and t < stim_off) * (mu0 - coh / 100.0 * mu1 + sigma*randn())", dt=stim_interval)
C_stimE1 = Synapses(stiminputE1, popE1, on_pre='s_AMPA_ext += gextE', name='C_stimE1')
C_stimE1.connect(j='i')
C_stimE2 = Synapses(stiminputE2, popE2, on_pre='s_AMPA_ext += gextE', name='C_stimE2')
C_stimE2.connect(j='i')
# -----------------------------------------------------------------------------------------------
# Run the simulation
# -----------------------------------------------------------------------------------------------
# Set initial conditions
popE.s_NMDA_tot = tau_NMDA_decay * 10 * Hz * 0.2
popI.s_NMDA_tot = tau_NMDA_decay * 10 * Hz * 0.2
popE.V = Vt - 2 * mV
popI.V = Vt - 2 * mV
# Record spikes of excitatory neurons in the decision encoding populations
SME1 = SpikeMonitor(popE1, record=True)
SME2 = SpikeMonitor(popE2, record=True)
# Record population activity
R1 = PopulationRateMonitor(popE1)
R2 = PopulationRateMonitor(popE2)
# Record input
E1 = StateMonitor(stiminputE1, 'rates', record=0, dt=1*ms)
E2 = StateMonitor(stiminputE2, 'rates', record=0, dt=1*ms)
# Run the simulation
run(runtime, report='stdout', profile=True)
print(profiling_summary())
# Show results
fig, axs = plt.subplots(4, 1, sharex=True, layout='constrained', gridspec_kw={'height_ratios': [2, 2, 2, 1]})
axs[0].plot(SME1.t / ms, SME1.i, '.', markersize=2, color='darkred')
axs[0].set(ylabel='population 1', ylim=(0, subN))
axs[1].plot(SME2.t / ms, SME2.i, '.', markersize=2, color='darkblue')
axs[1].set(ylabel='population 2', ylim=(0, subN))
axs[2].plot(R1.t / ms, R1.smooth_rate(window='flat', width=100 * ms) / Hz, color='darkred')
axs[2].plot(R2.t / ms, R2.smooth_rate(window='flat', width=100 * ms) / Hz, color='darkblue')
axs[2].set(ylabel='Firing rate (Hz)')
axs[3].plot(E1.t / ms, E1.rates[0] / Hz, color='darkred')
axs[3].plot(E2.t / ms, E2.rates[0] / Hz, color='darkblue')
axs[3].set(ylabel='Input (Hz)', xlabel='Time (ms)')
fig.align_ylabels(axs)
plt.show()