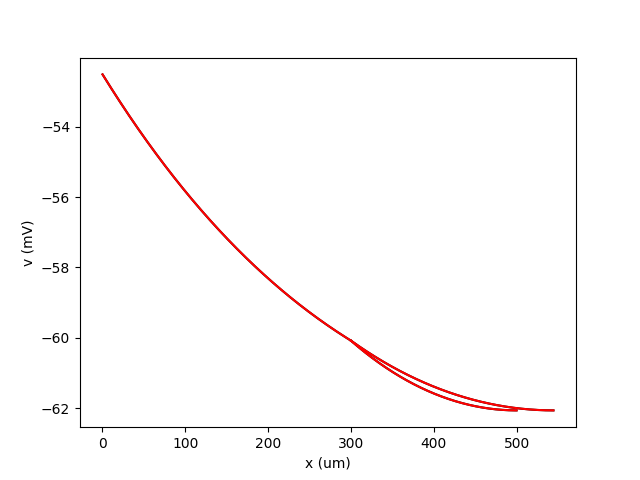
Example: rall
A cylinder plus two branches, with diameters according to Rall’s formula
from brian2 import *
defaultclock.dt = 0.01*ms
# Passive channels
gL = 1e-4*siemens/cm**2
EL = -70*mV
# Morphology
diameter = 1*um
length = 300*um
Cm = 1*uF/cm**2
Ri = 150*ohm*cm
N = 500
rm = 1 / (gL * pi * diameter) # membrane resistance per unit length
ra = (4 * Ri)/(pi * diameter**2) # axial resistance per unit length
la = sqrt(rm / ra) # space length
morpho = Cylinder(diameter=diameter, length=length, n=N)
d1 = 0.5*um
L1 = 200*um
rm = 1 / (gL * pi * d1) # membrane resistance per unit length
ra = (4 * Ri) / (pi * d1**2) # axial resistance per unit length
l1 = sqrt(rm / ra) # space length
morpho.L = Cylinder(diameter=d1, length=L1, n=N)
d2 = (diameter**1.5 - d1**1.5)**(1. / 1.5)
rm = 1/(gL * pi * d2) # membrane resistance per unit length
ra = (4 * Ri) / (pi * d2**2) # axial resistance per unit length
l2 = sqrt(rm / ra) # space length
L2 = (L1 / l1) * l2
morpho.R = Cylinder(diameter=d2, length=L2, n=N)
eqs='''
Im = gL * (EL-v) : amp/meter**2
I : amp (point current)
'''
neuron = SpatialNeuron(morphology=morpho, model=eqs, Cm=Cm, Ri=Ri,
method='exponential_euler')
neuron.v = EL
neuron.I[0] = 0.02*nA # injecting at the left end
run(100*ms, report='text')
plot(neuron.main.distance/um, neuron.main.v/mV, 'k')
plot(neuron.L.distance/um, neuron.L.v/mV, 'k')
plot(neuron.R.distance/um, neuron.R.v/mV, 'k')
# Theory
x = neuron.main.distance
ra = la * 4 * Ri/(pi * diameter**2)
l = length/la + L1/l1
theory = EL + ra*neuron.I[0]*cosh(l - x/la)/sinh(l)
plot(x/um, theory/mV, 'r')
x = neuron.L.distance
theory = (EL+ra*neuron.I[0]*cosh(l - neuron.main.distance[-1]/la -
(x - neuron.main.distance[-1])/l1)/sinh(l))
plot(x/um, theory/mV, 'r')
x = neuron.R.distance
theory = (EL+ra*neuron.I[0]*cosh(l - neuron.main.distance[-1]/la -
(x - neuron.main.distance[-1])/l2)/sinh(l))
plot(x/um, theory/mV, 'r')
xlabel('x (um)')
ylabel('v (mV)')
show()