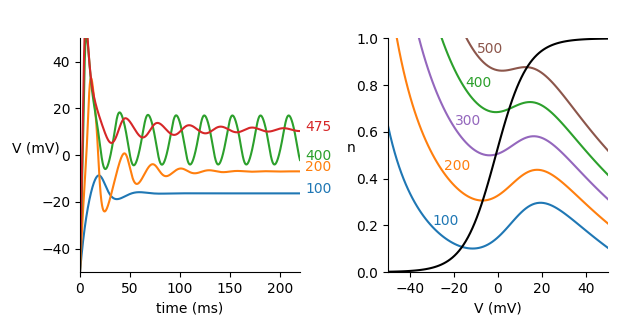
Example: Morris_Lecar_1981
Morris-Lecar model
Reproduces Fig. 9 of:
Catherine Morris and Harold Lecar. “Voltage Oscillations in the Barnacle Giant Muscle Fiber.” Biophysical Journal 35, no. 1 (1981): 193–213.
from brian2 import *
set_device('cpp_standalone')
defaultclock.dt = 0.01*ms
g_L = 2*mS
g_Ca = 4*mS
g_K = 8*mS
V_L = -50*mV
V_Ca = 100*mV
V_K = -70*mV
lambda_n__max = 1.0/(15*ms)
V_1 = 10*mV
V_2 = 15*mV # Note that Figure caption says -15 which seems to be a typo
V_3 = -1*mV
V_4 = 14.5*mV
C = 20*uF
# V,N-reduced system (Eq. 9 in article), note that the variables M and N (and lambda_N, etc.)
# have been renamed to m and n to better match the Hodgkin-Huxley convention, and because N has
# a reserved meaning in Brian (number of neurons)
eqs = '''
dV/dt = (-g_L*(V - V_L) - g_Ca*m_inf*(V - V_Ca) - g_K*n*(V - V_K) + I)/C : volt
dn/dt = lambda_n*(n_inf - n) : 1
m_inf = 0.5*(1 + tanh((V - V_1)/V_2)) : 1
n_inf = 0.5*(1 + tanh((V - V_3)/V_4)) : 1
lambda_n = lambda_n__max*cosh((V - V_3)/(2*V_4)) : Hz
I : amp
'''
neuron = NeuronGroup(17, eqs, method='exponential_euler')
neuron.I = (np.arange(17)*25+100)*uA
neuron.V = V_L
neuron.n = 'n_inf'
mon = StateMonitor(neuron, ['V', 'n'], record=True)
run_time = 220*ms
run(run_time)
fig, (ax1, ax2) = plt.subplots(1, 2, gridspec_kw={'right': 0.95, 'bottom': 0.15},
figsize=(6.4, 3.2))
fig.subplots_adjust(wspace=0.4)
for line_no, idx in enumerate([0, 4, 12, 15]):
color = 'C%d' % line_no
ax1.plot(mon.t/ms, mon.V[idx]/mV, color=color)
ax1.text(225, mon.V[idx][-1]/mV, '%.0f' % (neuron.I[idx]/uA), color=color)
ax1.set(xlim=(0, 220), ylim=(-50, 50), xlabel='time (ms)')
ax1.set_ylabel('V (mV)', rotation=0)
ax1.spines['right'].set_visible(False)
ax1.spines['top'].set_visible(False)
# dV/dt nullclines
V = linspace(-50, 50, 100)*mV
for line_no, (idx, color) in enumerate([(0, 'C0'), (4, 'C1'), (8, 'C4'), (12, 'C2'), (16, 'C5')]):
n_null = (g_L*(V - V_L) + g_Ca*0.5*(1 + tanh((V - V_1)/V_2))*(V - V_Ca) - neuron.I[idx])/(-g_K*(V - V_K))
ax2.plot(V/mV, n_null, color=color)
ax2.text(V[20+5*line_no]/mV, n_null[20+5*line_no]+0.01, '%.0f' % (neuron.I[idx]/uA), color=color)
# dn/dt nullcline
n_null = 0.5*(1 + tanh((V - V_3)/V_4))
ax2.plot(V/mV, n_null, color='k')
ax2.set(xlim=(-50, 50), ylim=(0, 1), xlabel='V (mV)')
ax2.set_ylabel('n', rotation=0)
ax2.spines['right'].set_visible(False)
ax2.spines['top'].set_visible(False)
plt.show()