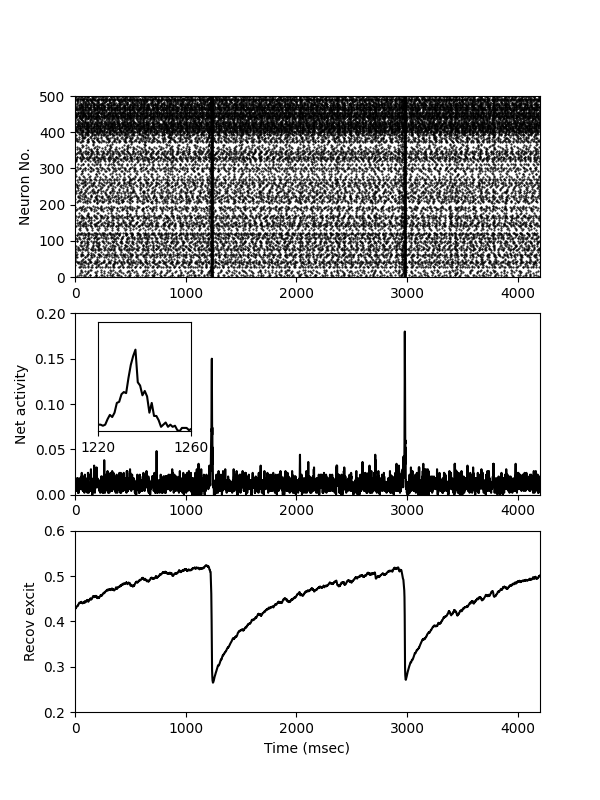
Example: Tsodyks_Uziel_Markram_2000
Fig. 1 from:
Synchrony Generation in Recurrent Networks with Frequency-Dependent Synapses The Journal of Neuroscience, 2000, Vol. 20 RC50
Implementation partially based on nest-2.0.0/examples/nest/tsodyks_shortterm_bursts.sli by Moritz Helias, 2006.
Sebastian Schmitt, 2022
import numpy as np
# set seed for reproducible figures
np.random.seed(5)
# for truncated normal
import scipy
from scipy import stats
import matplotlib.pyplot as plt
from brian2 import (
NeuronGroup,
Synapses,
SpikeGeneratorGroup,
SpikeMonitor,
StateMonitor,
)
from brian2 import ms, mV
from brian2 import run, defaultclock
def truncated_normal(loc, scale, bounds, size):
"""Normal distribution truncated within bounds
loc -- mean (“centre”) of the distribution
scale -- standard deviation (spread or “width”) of the distribution
bounds -- list of min and maximum
size -- number of samples
"""
bounds = np.array([bounds] * size)
s = scipy.stats.truncnorm.rvs(
(bounds[:, 0] - loc) / scale, (bounds[:, 1] - loc) / scale, loc=loc, scale=scale
)
return s
def get_population(name, N, tau_refrac):
"""Get population of neurons
name -- name of population
N -- number of neurons
tau_refrac -- refractory period
"""
neurons = NeuronGroup(
N,
"""
tau_mem : second
tau_refrac : second
v_reset : volt
v_thresh : volt
I_syn_ee_synapses : volt
I_syn_ei_synapses : volt
I_syn_ie_synapses : volt
I_syn_ii_synapses : volt
I_b : volt
dv/dt = -v/tau_mem + (I_syn_ee_synapses +
I_syn_ei_synapses +
I_syn_ie_synapses +
I_syn_ii_synapses)/tau_mem
+ I_b/tau_mem : volt (unless refractory)
""",
threshold="v>v_thresh",
reset="v=v_reset",
refractory=tau_refrac,
method="exact",
name=name,
)
v_thresh = 15 * mV
v_reset = 13.5 * mV
neurons.tau_mem = 30 * ms
neurons.v_thresh = v_thresh
neurons.v_reset = v_reset
# paper gives range of 0.05 mV but population bursts are not visible with that value
# -> increased to 1 mV range
neurons.I_b = (
np.random.uniform(v_thresh / mV - 0.5, v_thresh / mV + 0.5, size=N) * mV
)
return neurons
def get_synapses(name, source, target, tau_I, A, U, tau_rec, tau_facil=None):
"""Construct connections and retrieve synapses
name -- name of synapses
source -- source of connections
target -- target of connections
tau_I -- inactivation time constant
A -- absolute synaptic strength
U -- utilization of synaptic efficacy
tau_rec -- recovery time constant
tau_facil -- facilitation time constant (optional)
"""
synapses_eqs = """
A : volt
U : 1
tau_I : second
tau_rec : second
dx/dt = z/tau_rec : 1 (clock-driven) # recovered
dy/dt = -y/tau_I : 1 (clock-driven) # active
z = 1 - x - y : 1 # inactive
I_syn_{}_post = A*y : volt (summed)
""".format(
name
)
if tau_facil:
synapses_eqs += """
du/dt = -u/tau_facil : 1 (clock-driven)
tau_facil : second
"""
synapses_action = """
u += U*(1-u)
y += u*x # important: update y first
x += -u*x
"""
else:
synapses_action = """
y += U*x # important: update y first
x += -U*x
"""
synapses = Synapses(
source,
target,
model=synapses_eqs,
on_pre=synapses_action,
method="exact",
name=name,
)
synapses.connect(p=0.1)
N_syn = len(synapses)
synapses.tau_I = tau_I
A_min = min(0.2 * A, 2 * A)
A_max = max(0.2 * A, 2 * A)
synapses.A = (
truncated_normal(
A / mV, 0.5 * abs(A / mV), [A_min / mV, A_max / mV], size=N_syn
) * mV
)
assert not any(synapses.A < A_min)
assert not any(synapses.A > A_max)
U_mean, U_min, U_max = U
synapses.U = truncated_normal(U_mean, 0.5 * U_mean, [U_min, U_max], size=N_syn)
assert not any(synapses.U <= U_min)
assert not any(synapses.U > U_max)
tau_min = 5
synapses.tau_rec = (
truncated_normal(
tau_rec / ms, 0.5 * tau_rec / ms, [tau_min, np.inf], size=N_syn
) * ms
)
assert not any(synapses.tau_rec / ms <= tau_min)
if tau_facil:
synapses.tau_facil = (
truncated_normal(
tau_facil / ms, 0.5 * tau_facil / ms, [tau_min, np.inf], size=N_syn
) * ms
)
assert not any(synapses.tau_facil / ms <= tau_min)
# start fully recovered
synapses.x = 1
return synapses
# configure neuron populations
exc_neurons = get_population("exc_neurons", N=400, tau_refrac=3 * ms)
inh_neurons = get_population("inh_neurons", N=100, tau_refrac=2 * ms)
# configure synapses
ee_synapses = get_synapses(
"ee_synapses",
exc_neurons,
exc_neurons,
tau_I=3 * ms,
A=1.8 * mV,
U=[0.5, 0.1, 0.9],
tau_rec=800 * ms,
)
ei_synapses = get_synapses(
"ei_synapses",
exc_neurons,
inh_neurons,
tau_I=3 * ms,
A=7.2 * mV,
U=[0.04, 0.001, 0.07],
tau_rec=100 * ms,
tau_facil=1000 * ms,
)
ie_synapses = get_synapses(
"ie_synapses",
inh_neurons,
exc_neurons,
tau_I=3 * ms,
A=-5.4 * mV,
U=[0.5, 0.1, 0.9],
tau_rec=800 * ms,
)
ii_synapses = get_synapses(
"ii_synapses",
inh_neurons,
inh_neurons,
tau_I=3 * ms,
A=-7.2 * mV,
U=[0.04, 0.001, 0.07],
tau_rec=100 * ms,
tau_facil=1000 * ms,
)
# run for burnin time to settle network activity
defaultclock.dt = 1 * ms
burnin = 900
run(burnin * ms)
# record from now on
spike_monitor_exc = SpikeMonitor(exc_neurons)
spike_monitor_inh = SpikeMonitor(inh_neurons)
state_monitor_ee = StateMonitor(ee_synapses, ["x"], record=True)
duration = 4200
run(duration * ms, report="text")
# plots
fig, axes = plt.subplots(3, figsize=(6, 8), sharex=True)
# raster plot
axes[0].plot(spike_monitor_exc.t / ms, spike_monitor_exc.i, ".k", ms=1)
axes[0].plot(spike_monitor_inh.t / ms, spike_monitor_inh.i + len(exc_neurons), ".k", ms=1)
axes[0].set_ylabel("Neuron No.")
axes[0].set_ylim(0, len(exc_neurons) + len(inh_neurons))
# network activity
net_activity = np.histogram(
np.concatenate(
list(spike_monitor_exc.spike_trains().values())
+ list(spike_monitor_inh.spike_trains().values())
) / ms,
bins=np.arange(burnin, duration + burnin, 1))[0] / (len(exc_neurons) + len(inh_neurons))
axes[1].plot(np.arange(0, len(net_activity)) + burnin, net_activity, "k")
net_activity_min = 0
net_activity_max = 0.2
axes[1].set_ylim(net_activity_min, net_activity_max)
axes[1].set_ylabel("Net activity")
# network activity inset
axins = axes[1].inset_axes([0.05, 0.35, 0.2, 0.6])
axins.plot(np.arange(0, len(net_activity)) + burnin, net_activity, "k")
inset_min = 1220
inset_max = 1260
axins.set_xlim(inset_min + burnin, inset_max + burnin)
axins.set_ylim(net_activity_min, net_activity_max)
axins.set_xticks([inset_min + burnin, inset_max + burnin])
axins.set_xticklabels([inset_min, inset_max])
axins.set_yticks([])
# recovered synaptic partition
axes[2].plot(
state_monitor_ee.t / ms, np.mean(state_monitor_ee.x, axis=0), "k", label="x"
)
axes[2].set_ylim(0.2, 0.6)
axes[2].set_xlabel("Time (msec)")
axes[2].set_ylabel("Recov excit")
axes[2].set_xlim(burnin, duration + burnin)
xtickstep = 1000
axes[2].set_xticks(np.arange(burnin, duration + burnin, xtickstep))
axes[2].set_xticklabels(map(str, range(0, duration, xtickstep)))
axes[0].xaxis.set_tick_params(which="both", labelbottom=True)
axes[1].xaxis.set_tick_params(which="both", labelbottom=True)
plt.show()