Example: Nicola_Clopath_2017
FORCE training of a Leaky IF model to mimic a sinusoid (5 Hz) oscillator
Nicola, W., Clopath, C. Supervised learning in spiking neural networks with FORCE training Nat Commun 8, 2208 (2017)
https://doi.org/10.1038/s41467-017-01827-3
Based on https://github.com/ModelDBRepository/190565/blob/master/CODE%20FOR%20FIGURE%202/LIFFORCESINE.m
Sebastian Schmitt, 2022
from brian2 import NeuronGroup, Synapses, StateMonitor, SpikeMonitor
from brian2 import run, defaultclock, network_operation
from brian2 import ms, second, Hz
import matplotlib.pyplot as plt
from matplotlib.ticker import MaxNLocator
import numpy as np
# set seed for reproducible figures
np.random.seed(1)
# decay time of synaptic kernal
td = 20*ms
# rise time of synaptic kernal
tr = 2*ms
# membrane time constant
tm = 10*ms
# refractory period
tref = 2*ms
# reset potential
vreset = -65
# peak/threshold potential
vpeak = -40
# bias
BIAS = vpeak
# integration time step
defaultclock.dt = 0.05*ms
# total duration of simulation
T = 15*second
# start of training
imin = 5*second
# end of training
icrit = 10*second
# interval of training
step = 2.5*ms
# feedback scale factor
Q = 10
# neuron-to-neuron connection scale factor
G = 0.04
# connection probability
p = 0.1
# number of neurons
N = 2000
# correlation weight matrix for RLMS
alpha = defaultclock.dt/second*0.1
Pinv = np.eye(N)*alpha
# Sinusoid oscillator
def zx(t):
freq = 5*Hz
return np.sin(2*np.pi*freq*t)
neurons = NeuronGroup(N,
"""
dv/dt = (-v + BIAS + IPSC + E*z)/tm: 1 (unless refractory)
dIPSC/dt = -IPSC/tr + h : 1
dh/dt = -h/td : 1/second
dr/dt = -r/tr + hr : 1
dhr/dt = -hr/td : 1/second
BPhi : 1
z : 1 (shared)
E : 1
""",
method="euler",
threshold="v>=vpeak",
reset="v=vreset; hr += 1/(tr*td)*second",
refractory=tref)
# fixed feedback weights
neurons.E = (2*np.random.uniform(size=N)-1)*Q
# initial membrane voltage
neurons.v = vreset + np.random.uniform(size=N)*(30-vreset)
synapses = Synapses(neurons, neurons, "w : second", on_pre="h += w/(tr*td)")
synapses.connect()
omega = G*(np.random.normal(size=(N,N))*(np.random.uniform(size=(N,N))<p))/(np.sqrt(N)*p)
synapses.w = omega.flatten()*second
spikemon = SpikeMonitor(neurons[:20])
statemon_BPhi = StateMonitor(neurons, "BPhi", record=range(10))
statemon_z = StateMonitor(neurons, "z", record=[0])
# linear readout
@network_operation(dt=defaultclock.dt)
def readout(t):
neurons.z = np.dot(neurons.BPhi, neurons.r)
# FORCE training
@network_operation(dt=step)
def train(t):
global Pinv
if t > imin and t < icrit:
cd = Pinv@neurons.r
err = neurons.z - zx(t)
neurons.BPhi -= cd*err
Pinv -= np.outer(cd,cd)/( 1 + np.dot(neurons.r, cd))
run(T, report="text")
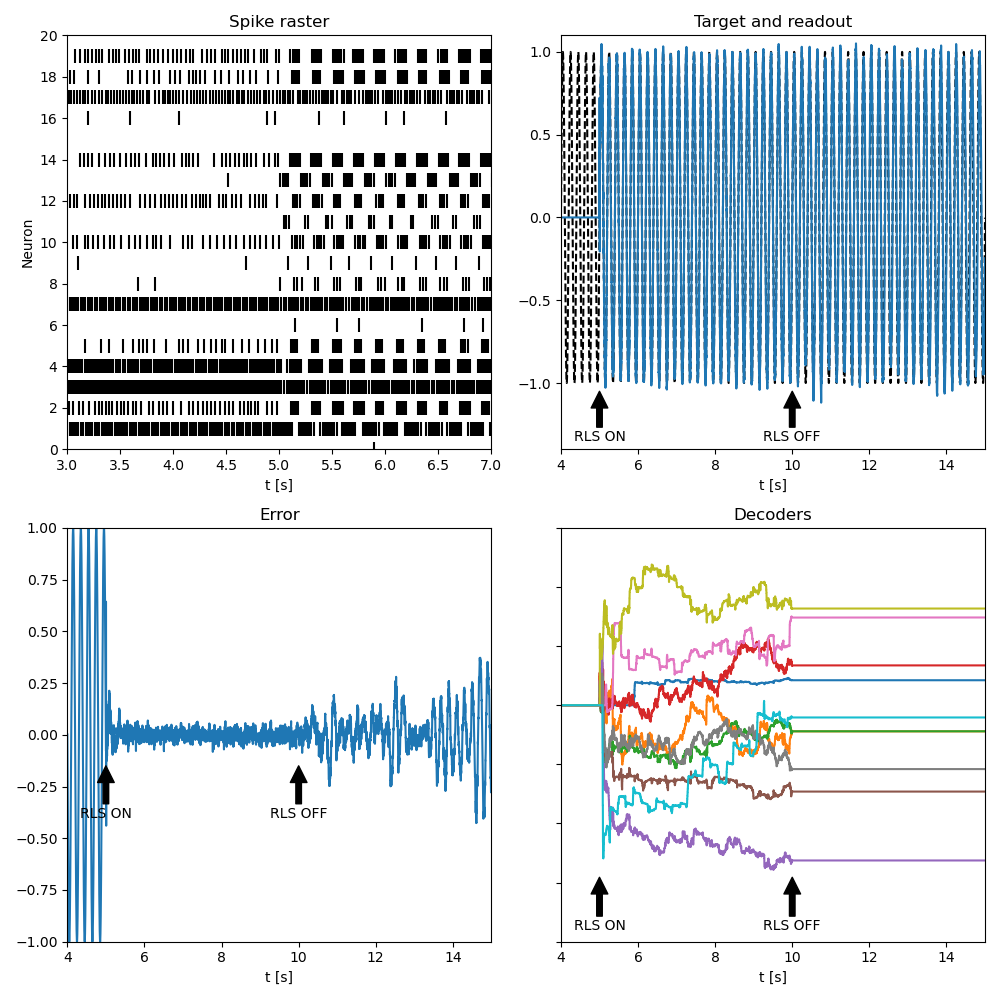
fig, axes = plt.subplots(2,2, figsize=(10,10))
axes = axes.flatten()
axes[0].set_title("Spike raster")
axes[0].scatter(spikemon.t/second,spikemon.i, marker='|', linestyle="None", color="black", s=100)
axes[0].set_xlim((imin-2*second)/second, imin/second+2)
axes[0].set_ylim(0, len(spikemon.source))
axes[0].set_xlabel("t [s]")
axes[0].set_ylabel("Neuron")
axes[0].yaxis.set_major_locator(MaxNLocator(integer=True))
axes[1].plot(statemon_z.t/second, zx(statemon_z.t), linestyle='--', color='k')
axes[1].plot(statemon_z.t/second,statemon_z.z[0])
axes[1].set_title("Target and readout")
axes[1].annotate('RLS ON', xy=(imin/second, -1.05), xytext=(imin/second, -1.35),
arrowprops=dict(facecolor='black', shrink=1), ha="center")
axes[1].annotate('RLS OFF', xy=(icrit/second, -1.05), xytext=(icrit/second, -1.35),
arrowprops=dict(facecolor='black', shrink=1), ha="center")
axes[1].set_xlabel("t [s]")
axes[1].set_xlim((imin-1*second)/second, T/second)
axes[1].set_ylim(-1.4,1.1)
axes[2].set_title("Error")
axes[2].plot(statemon_z.t/second, statemon_z.z[0] - zx(statemon_z.t))
axes[2].annotate('RLS ON', xy=(imin/second, -0.15), xytext=(imin/second, -0.4),
arrowprops=dict(facecolor='black', shrink=1), ha="center")
axes[2].annotate('RLS OFF', xy=(icrit/second, -0.15), xytext=(icrit/second, -0.4),
arrowprops=dict(facecolor='black', shrink=1), ha="center")
axes[2].set_xlabel("t [s]")
axes[2].set_xlim((imin-1*second)/second, T/second)
axes[2].set_ylim(-1,1)
axes[3].set_title("Decoders")
for j in range(len(statemon_BPhi.record)):
axes[3].plot(statemon_BPhi.t/second,statemon_BPhi.BPhi[j])
axes[3].set_xlim((imin-1*second)/second, T/second)
axes[3].set_xlabel("t [s]")
axes[3].set_ylim(-0.00020, 0.00015)
axes[3].set_yticklabels([])
axes[3].annotate('RLS ON', xy=(imin/second, -0.0001455), xytext=(imin/second, -0.00019),
arrowprops=dict(facecolor='black', shrink=1), ha="center")
axes[3].annotate('RLS OFF', xy=(icrit/second, -0.0001455), xytext=(icrit/second, -0.00019),
arrowprops=dict(facecolor='black', shrink=1), ha="center")
fig.tight_layout()