Example: custom_events¶
Example demonstrating the use of custom events.
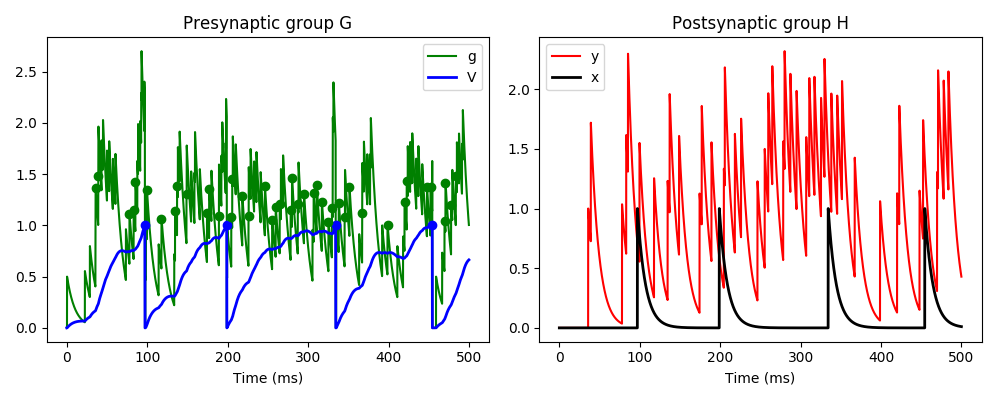
Here we have three neurons, the first is Poisson spiking and connects to neuron G,
which in turn connects to neuron H. Neuron G has two variables v and g, and the
incoming Poisson spikes cause an instantaneous increase in variable g. g decays
rapidly, and in turn causes a slow increase in v. If v crosses a threshold, it
causes a standard spike and reset. If g crosses a threshold, it causes a custom
event gspike, and if it returns below that threshold it causes a custom
event end_gspike. The standard spike event when v crosses a threshold
causes an instantaneous increase in variable x in neuron H (which happens
through the standard pre pathway in the synapses), and the gspike
event causes an increase in variable y (which happens through the custom
pathway gpath).
from brian2 import *
# Input Poisson spikes
inp = PoissonGroup(1, rates=250*Hz)
# First group G
eqs_G = '''
dv/dt = (g-v)/(50*ms) : 1
dg/dt = -g/(10*ms) : 1
allow_gspike : boolean
'''
G = NeuronGroup(1, eqs_G, threshold='v>1',
reset='v = 0; g = 0; allow_gspike = True;',
events={'gspike': 'g>1 and allow_gspike',
'end_gspike': 'g<1 and not allow_gspike'})
G.run_on_event('gspike', 'allow_gspike = False')
G.run_on_event('end_gspike', 'allow_gspike = True')
# Second group H
eqs_H = '''
dx/dt = -x/(10*ms) : 1
dy/dt = -y/(10*ms) : 1
'''
H = NeuronGroup(1, eqs_H)
# Synapses from input Poisson group to G
Sin = Synapses(inp, G, on_pre='g += 0.5')
Sin.connect()
# Synapses from G to H
S = Synapses(G, H,
on_pre={'pre': 'x += 1',
'gpath': 'y += 1'},
on_event={'pre': 'spike',
'gpath': 'gspike'})
S.connect()
# Monitors
Mstate = StateMonitor(G, ('v', 'g'), record=True)
Mgspike = EventMonitor(G, 'gspike', 'g')
Mspike = SpikeMonitor(G, 'v')
MHstate = StateMonitor(H, ('x', 'y'), record=True)
# Initialise and run
G.allow_gspike = True
run(500*ms)
# Plot
figure(figsize=(10, 4))
subplot(121)
plot(Mstate.t/ms, Mstate.g[0], '-g', label='g')
plot(Mstate.t/ms, Mstate.v[0], '-b', lw=2, label='V')
plot(Mspike.t/ms, Mspike.v, 'ob', label='_nolegend_')
plot(Mgspike.t/ms, Mgspike.g, 'og', label='_nolegend_')
xlabel('Time (ms)')
title('Presynaptic group G')
legend(loc='best')
subplot(122)
plot(MHstate.t/ms, MHstate.y[0], '-r', label='y')
plot(MHstate.t/ms, MHstate.x[0], '-k', lw=2, label='x')
xlabel('Time (ms)')
title('Postsynaptic group H')
legend(loc='best')
tight_layout()
show()