Example: Clopath_et_al_2010_homeostasis¶
This code contains an adapted version of the voltage-dependent triplet STDP rule from: Clopath et al., Connectivity reflects coding: a model of voltage-based STDP with homeostasis, Nature Neuroscience, 2010 (http://dx.doi.org/10.1038/nn.2479)
The plasticity rule is adapted for a leaky integrate & fire model in Brian2.
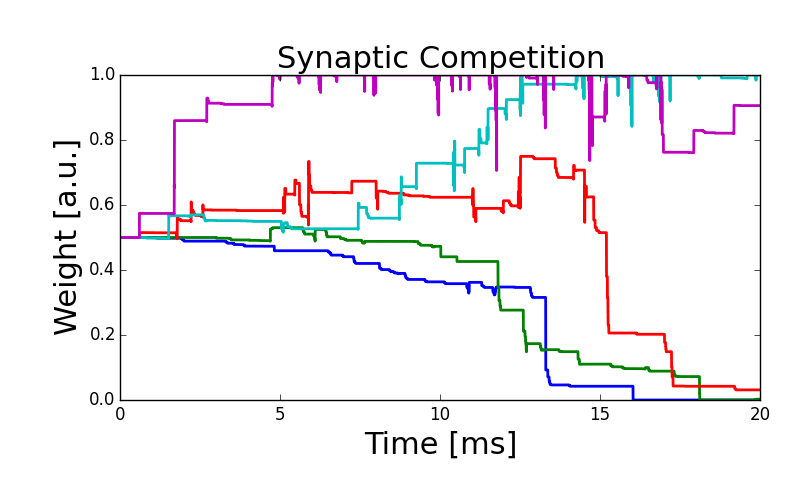
As an illustration of the rule, we simulate the competition between inputs projecting on a downstream neuron
We kindly ask to cite the article when using the model presented below.
This code was written by Jacopo Bono, 12/2015
from brian2 import *
################################################################################
# PLASTICITY MODEL
################################################################################
#### Plasticity Parameters
V_rest = -70.*mV # resting potential
V_thresh = -55.*mV # spiking threshold
Theta_low = V_rest # depolarization threshold for plasticity
x_reset = 1. # spike trace reset value
taux = 15.*ms # spike trace time constant
A_LTD = 1.5e-4 # depression amplitude
A_LTP = 1.5e-2 # potentiation amplitude
tau_lowpass1 = 40*ms # timeconstant for low-pass filtered voltage
tau_lowpass2 = 30*ms # timeconstant for low-pass filtered voltage
tau_homeo = 1000*ms # homeostatic timeconstant
v_target = 12*mV**2 # target depolarisation
#### Plasticity Equations
# equations executed at every timestep
Syn_model = ('''
w_ampa:1 # synaptic weight (ampa synapse)
''')
# equations executed only when a presynaptic spike occurs
Pre_eq = ('''
g_ampa_post += w_ampa*ampa_max_cond # increment synaptic conductance
A_LTD_u = A_LTD*(v_homeo**2/v_target) # metaplasticity
w_minus = A_LTD_u*(v_lowpass1_post/mV - Theta_low/mV)*(v_lowpass1_post/mV - Theta_low/mV > 0) # synaptic depression
w_ampa = clip(w_ampa-w_minus,0,w_max) # hard bounds
''' )
# equations executed only when a postsynaptic spike occurs
Post_eq = ('''
v_lowpass1 += 10*mV # mimics the depolarisation effect due to a spike
v_lowpass2 += 10*mV # mimics the depolarisation effect due to a spike
v_homeo += 0.1*mV # mimics the depolarisation effect due to a spike
w_plus = A_LTP*x_trace_pre*(v_lowpass2_post/mV - Theta_low/mV)*(v_lowpass2_post/mV - Theta_low/mV > 0) # synaptic potentiation
w_ampa = clip(w_ampa+w_plus,0,w_max) # hard bounds
''' )
################################################################################
# I&F Parameters and equations
################################################################################
#### Neuron parameters
gleak = 30.*nS # leak conductance
C = 300.*pF # membrane capacitance
tau_AMPA = 2.*ms # AMPA synaptic timeconstant
E_AMPA = 0.*mV # reversal potential AMPA
ampa_max_cond = 5.e-8*siemens # Ampa maximal conductance
w_max = 1. # maximal ampa weight
#### Neuron Equations
# We connect 10 presynaptic neurons to 1 downstream neuron
# downstream neuron
eqs_neurons = '''
dv/dt = (gleak*(V_rest-v) + I_ext + I_syn)/C: volt # voltage
dv_lowpass1/dt = (v-v_lowpass1)/tau_lowpass1 : volt # low-pass filter of the voltage
dv_lowpass2/dt = (v-v_lowpass2)/tau_lowpass2 : volt # low-pass filter of the voltage
dv_homeo/dt = (v-V_rest-v_homeo)/tau_homeo : volt # low-pass filter of the voltage
I_ext : amp # external current
I_syn = g_ampa*(E_AMPA-v): amp # synaptic current
dg_ampa/dt = -g_ampa/tau_AMPA : siemens # synaptic conductance
dx_trace/dt = -x_trace/taux :1 # spike trace
'''
# input neurons
eqs_inputs = '''
dv/dt = gleak*(V_rest-v)/C: volt # voltage
dx_trace/dt = -x_trace/taux :1 # spike trace
rates : Hz # input rates
selected_index : integer (shared) # active neuron
'''
################################################################################
# Simulation
################################################################################
#### Parameters
defaultclock.dt = 500.*us # timestep
Nr_neurons = 1 # Number of downstream neurons
Nr_inputs = 5 # Number of input neurons
input_rate = 35*Hz # Rates
init_weight = 0.5 # initial synaptic weight
final_t = 20.*second # end of simulation
input_time = 100.*ms # duration of an input
#### Create neuron objects
Nrn_downstream = NeuronGroup(Nr_neurons, eqs_neurons, threshold='v>V_thresh',
reset='v=V_rest;x_trace+=x_reset/(taux/ms)',
method='euler')
Nrns_input = NeuronGroup(Nr_inputs, eqs_inputs, threshold='rand()<rates*dt',
reset='v=V_rest;x_trace+=x_reset/(taux/ms)',
method='linear')
#### create Synapses
Syn = Synapses(Nrns_input, Nrn_downstream,
model=Syn_model,
on_pre=Pre_eq,
on_post=Post_eq
)
Syn.connect(i=numpy.arange(Nr_inputs), j=0)
#### Monitors and storage
W_evolution = StateMonitor(Syn, 'w_ampa', record=True)
#### Run
# Initial values
Nrn_downstream.v = V_rest
Nrn_downstream.v_lowpass1 = V_rest
Nrn_downstream.v_lowpass2 = V_rest
Nrn_downstream.v_homeo = 0
Nrn_downstream.I_ext = 0.*amp
Nrn_downstream.x_trace = 0.
Nrns_input.v = V_rest
Nrns_input.x_trace = 0.
Syn.w_ampa = init_weight
# Switch on a different input every 100ms
Nrns_input.run_regularly('''
selected_index = int(floor(rand()*Nr_inputs))
rates = input_rate * int(selected_index == i) # All rates are zero except for the selected neuron
''', dt=input_time)
run(final_t, report='text')
################################################################################
# Plots
################################################################################
stitle = 'Synaptic Competition'
fig = figure(figsize=(8, 5))
for kk in range(Nr_inputs):
plt.plot(W_evolution.t, W_evolution.w_ampa[kk], '-', linewidth=2)
xlabel('Time [ms]', fontsize=22)
ylabel('Weight [a.u.]', fontsize=22)
plt.subplots_adjust(bottom=0.2, left=0.15, right=0.95, top=0.85)
title(stitle, fontsize=22)
plt.show()