Example: Brette_Guigon_2003¶
Reliability of spike timing¶
Adapted from Fig. 10D,E of
Brette R and E Guigon (2003). Reliability of Spike Timing Is a General Property of Spiking Model Neurons. Neural Computation 15, 279-308.
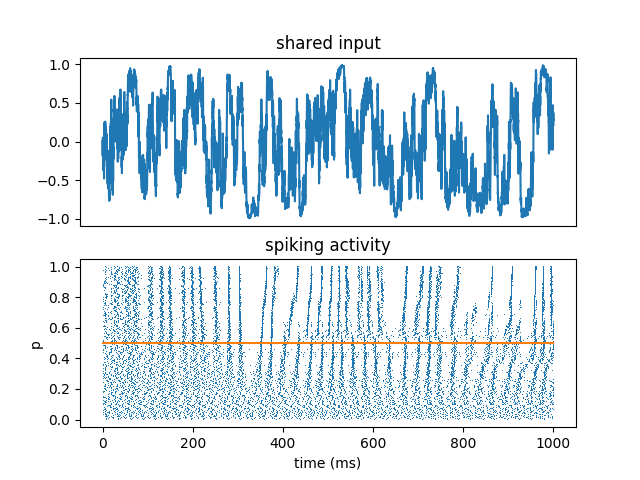
This shows that reliability of spike timing is a generic property of spiking neurons, even those that are not leaky. This is a non-physiological model which can be leaky or anti-leaky depending on the sign of the input I.
All neurons receive the same fluctuating input, scaled by a parameter p that varies across neurons. This shows:
reproducibility of spike timing
robustness with respect to deterministic changes (parameter)
increased reproducibility in the fluctuation-driven regime (input crosses the threshold)
from brian2 import *
N = 500
tau = 33*ms
taux = 20*ms
sigma = 0.02
eqs_input = '''
dx/dt = -x/taux + (2/taux)**.5*xi : 1
'''
eqs = '''
dv/dt = (v*I + 1)/tau + sigma*(2/tau)**.5*xi : 1
I = 0.5 + 3*p*B : 1
B = 2./(1 + exp(-2*x)) - 1 : 1 (shared)
p : 1
x : 1 (linked)
'''
input = NeuronGroup(1, eqs_input, method='euler')
neurons = NeuronGroup(N, eqs, threshold='v>1', reset='v=0', method='euler')
neurons.p = '1.0*i/N'
neurons.v = 'rand()'
neurons.x = linked_var(input, 'x')
M = StateMonitor(neurons, 'B', record=0)
S = SpikeMonitor(neurons)
run(1000*ms, report='text')
subplot(211) # The input
plot(M.t/ms, M[0].B)
xticks([])
title('shared input')
subplot(212)
plot(S.t/ms, neurons.p[S.i], ',')
plot([0, 1000], [.5, .5], color='C1')
xlabel('time (ms)')
ylabel('p')
title('spiking activity')
show()